What is PacBio SMRT sequencing?
Image credit: Philip Mynott/Wellcome Sanger Institute

A type of third generation sequencing, Single-Molecule Sequencing in Real Time (SMRT) made it possible to sequence the DNA from single cells for the first time.
- In 2011, PacBio released Single-Molecule Sequencing in Real Time (SMRT) – a new type of sequencing technique.
- Unlike previous techniques, SMRT could sequence individual molecules and individual cells for the first time.
- It was used to sequence the E coli genome with 99.9999% accuracy.
Key terms
DNA
(deoxyribonucleic acid) A molecule that carries the genetic information necessary to build and maintain an organism.
DNA sequencing
The process of determining the order of bases in a section of DNA.
What is PacBio Single-Molecule Sequencing in Real Time (SMRT)?
- SMRT is a third generation sequencing technology, developed by Pacific Biosciences (PacBio).
- Between 2011 and 2018, the technology evolved from being able to read fragments of around 1,000 bases to 30,000 bases in one go – known as a long read. Comparatively, earlier techniques like 454 sequencing could only read up to 400 bases.
- Initially, its speed and long read ability meant lower accuracy than early methods. However, because it is so fast and any errors are random, it’s possible to repeat the experiment and achieve up to 99.9999% accuracy.
How does SMRT work?
- SMRT involves a single-stranded molecule of DNA, bound to a DNA polymerase enzyme.
- The bound pair enter a sequencing chamber, called a flow cell.
- Like in Sanger sequencing, the DNA polymerase adds complementary, fluorescently labelled bases to the DNA strand.
- As each labelled base is added, the fluorescent colour of the base is recorded before the fluorescent label is cut off. The next base in the DNA chain can then be added and recorded.
- This is translated into a sequence by an algorithm.
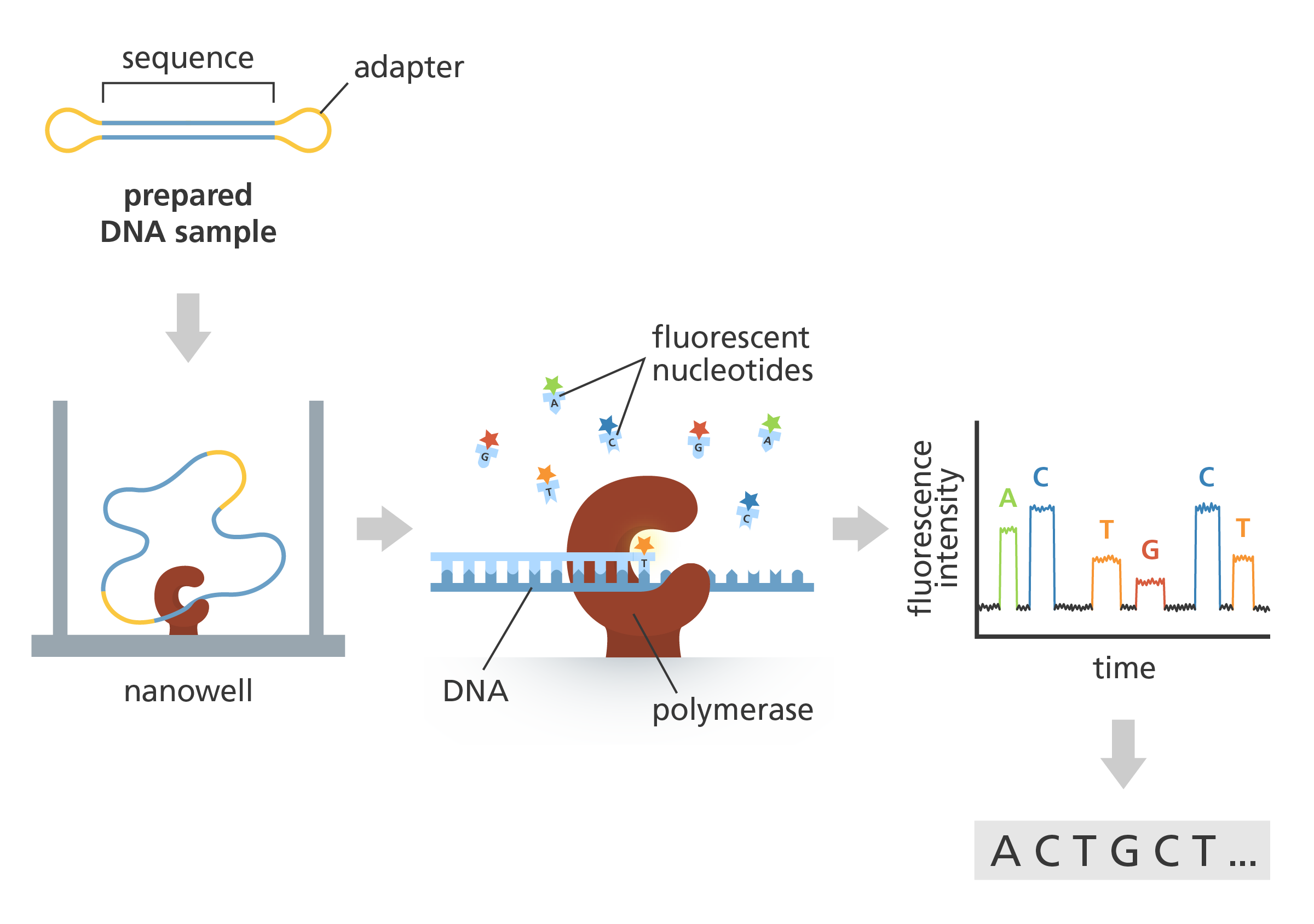
Single-Molecule Sequencing in Real Time (SMRT). A prepared DNA sample is bound to the polymerase (brown) via an adapter (yellow) and flown onto the flow cell. A DNA polymerase is connected to the bottom of each nanowell and a mix of fluorescently labelled nucleotides is added. The incorporation of each fluorescent nucleotide leads to a burst of light captured in the raw video data. Image credit: Laura Olivares Boldú, Wellcome Connecting Science
What are the benefits of SMRT?
- Cost and speed: SMRT is very efficient and uses fewer expensive chemicals than other techniques.
- Sensitivity: It is incredibly sensitive and fast, enabling scientists to effectively ‘eavesdrop’ on the DNA polymerase as it makes a strand of DNA, and read the sequence in real-time.
- Length: It can generate very long sequences (up to 30,000 bases long) from single molecules of DNA. This makes it easier to assemble full genomes.
- New sequences: Unlike Sanger sequencing, it doesn’t require a primer – so it’s possible to sequence species that have never been investigated before. This is known as de novo sequencing.
- Accuracy: Using SMRT, scientists sequenced the genome of the bacteria E coli to an accuracy of 99.9999%.