What is Oxford Nanopore Technologies (ONT) sequencing?
Image credit: Oxford Nanopore Technologies

Oxford Nanopore Technologies developed third generation sequencers that are portable, able to sequence DNA in remote locations and produce ultra-long reads.
- In 2015, the first nanopore sequencing device was released by Oxford Nanopore Technologies – an entirely new way to sequence DNA.
- Unlike previous techniques, which were based on DNA replication, ONT doesn’t use any DNA polymerases. Instead, it’s based on a small barrel-shaped protein in a membrane, called a nanopore, and measures changes to electrical current.
- It was used in 2015 during an Ebola outbreak to help with genomic surveillance.
Key terms
DNA
(deoxyribonucleic acid) A molecule that carries the genetic information necessary to build and maintain an organism.
DNA sequencing
The process of determining the order of bases in a section of DNA.
What is nanopore sequencing?
- Nanopore sequencing is a type of third generation sequencing technology from ONT.
- Unlike all previous sequencing technologies, ONT doesn’t use any DNA polymerases.
- Instead, it uses a barrel-shaped protein called CsgG – naturally found as a ‘pore’ in a cell membrane, regulating which molecules can enter or leave a cell. This is called a nanopore.
- CsgG has a diameter of 1 nanometre at its smallest point – just big enough to allow a single strand of DNA through.
How does nanopore sequencing work?
- In nanopore sequencing, the CsgG nanopore is embedded into an artificial membrane inside a sequencing chamber. When an electrical current is applied across the membrane, the current has to flow through the nanopore.
- DNA is threaded through the nanopore by an enzyme - a molecular motor - and it obstructs the current flowing through the nanopore.
- The four bases of the DNA (A, T, C and G) are of different shape and size, so cause characteristic variations in the current.
- These variations are measured by an electronic chip. An algorithm converts the data into a sequence which can then be read.
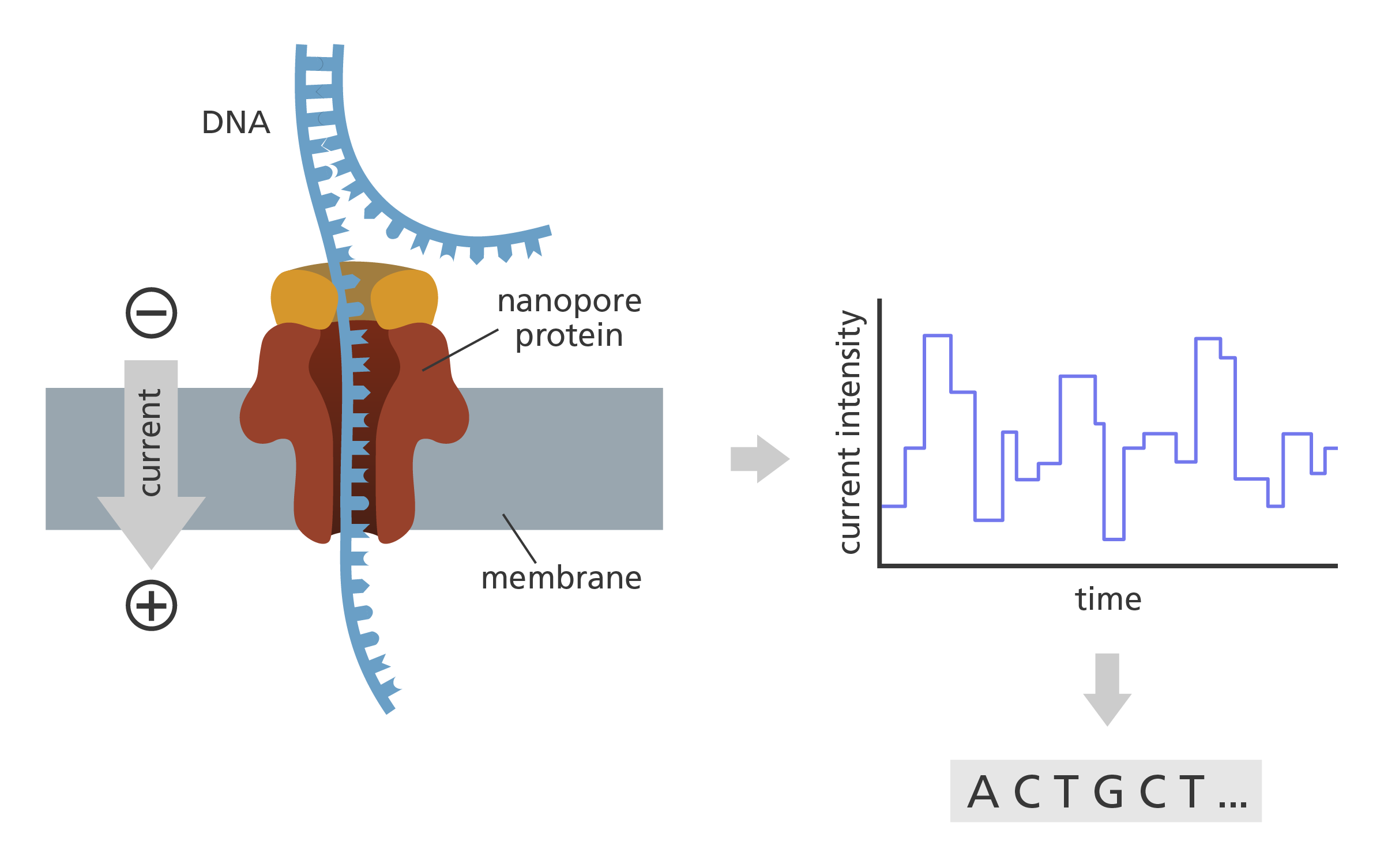
Oxford Nanopore Technologies. Nanopore proteins are embedded into an artificial membrane inside the sequencing flow cell. The obstruction of a nanopore by a DNA fragment leads to a change in the current that is measured continuously by an electronics chip integrated within the flow cell. Image credit: Laura Olivares Boldú, Wellcome Connecting Science
What are the benefits of nanopore sequencing?
- It can generate both very short (20 bases) and ultra-long reads (up to several millions of base pairs) making it much easier to sequence an entire genome.
- The instrument is very small – depending on the model, between the size of a phone and a microwave. Comparatively, Illumina and PacBio sequencers are closer in size to fridge-freezers. ONT’s small size makes the technology more accessible outside of the traditional laboratory setting, requiring only a laptop to run it.
- For example, the smallest ONT sequencer - the MinION - has been used onboard the International Space Station. It has also been used in low-income countries, including during the Ebola pandemic in 2015 – when it read the genomes of Ebola viruses from 14 patients in just 48 hours.
- The biggest ONT sequencer - the PromethION - can be used for human genome sequencing and was vital for filling in the gaps when the human genome was finally complete in 2021.

MinION sequencing device being used in Guinea during the 2013-2016 Ebola outbreak
Image credit: Tommy Trenchard / European Mobile Laboratories