What is bioinformatics and how do we use it?
Image credit: Greg Moss / Wellcome Sanger Institute

Bioinformatics is the science of both storing lots of complex biological data, and of analysing it to find new insights, which we use in many different ways.
- Bioinformatics is fundamental to much biological research and involves biologists who learn programming, or computer programmers, mathematicians or database managers who learn the foundations of biology.
- Bioinformatics enables us to handle the huge amounts of data involved and make sense of them.
What is bioinformatics?
- Modern science isn't simply about publishing one set of results and hoping other researchers read it. It's about linking everything that is out there, to provide new insights that we can only spot if we can see the big picture. Bioinformatics lets us bring together the data from lots of experiments in one place, so we can ask those big questions – and find the answers.
- Bioinformatics involves processing, storing and analysing biological data. This might include:
- Creating databases to store experimental data
- Predicting the way that proteins fold up
- Modelling how all the chemical reactions in a cell interact with each other
- Bioinformatics is a broad field and needs a diverse range of people with diverse skill sets. Programmers to write the computer programs to analyse all this data, database administrators to organise storing it all, biological scientists and statisticians to analyse the data, and web designers to produce sites and apps that scientists can use to search all this data.
What type of biological data can be used in bioinformatics?
Transcriptomics: the study of the transcriptome, the full set of RNA transcripts in a cell.
- Genes aren’t constantly active. They can be turned on and off by proteins and chemical messengers. A gene that is turned on, or expressed, will be used to produce RNA, which is then used as the assembly instructions for a protein.
- For example, your body makes haemoglobin to carry oxygen in red blood cells, but it’s not needed in white blood cells. We would therefore find RNA linked to haemoglobin production in the tissues that make red blood cells but not in the tissues where white blood cells are produced.
- Scientists can use RNA sequencing to compare gene expression in different cell types, for example between healthy and diseased cells.
Proteomics: the study of the complete set of proteins in a cell or system.
- Genes provide the information our cells use to make proteins, which are the machinery of the cell.
- Scientists can analyse a tissue sample and see what proteins can be found in it.
Phenomics: the study of phenotypes at a genome-wide scale.
- A phenotype is the way scientists describe something that can be measured about a person. A phenotype might be 'risk of diabetes' or 'eye colour'.
- Bioinformatics lets us look for possible links between our DNA and a phenotype.
Chemoinformatics: the computational analysis of chemical and biochemical data.
- Drug research generates lots of experimental data.
- Big databases of drug information can help scientists develop new drugs, by providing examples of chemicals that target a certain protein.
How can we use bioinformatics to answer questions in genomics?
It’s possible to start with any of the types of bioinformatics data shown above, depending on what question a lab wants to answer. There are two main approaches:
Starting from the genome:
- Tools like HumanMine and OpenTargets allow scientists to start from a gene and see what proteins it is the blueprint for.
- From there, they look at where the proteins are found in the body, and what diseases are linked to them.
Starting from the population:
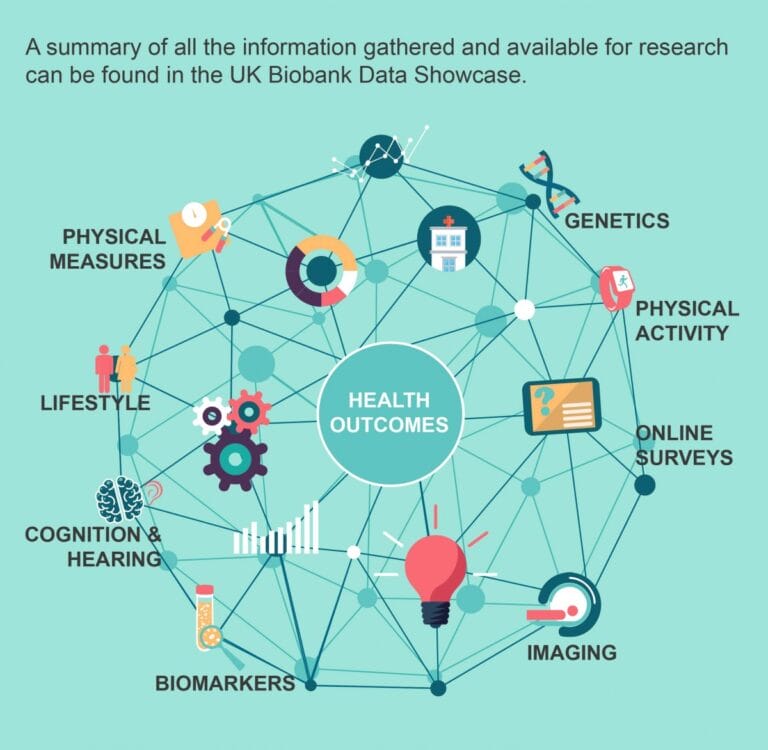
- Health researchers start with a large-scale study of volunteers who agree to share their phenotype measurements and a genetic sample.
- This population data lets researchers see if a phenotype is linked to a disease, or locate a gene that might be influencing the phenotype.
- Volunteers in big studies like UK Biobank mean that there is ready-made bioinformatics data available to researchers who apply for permission to use it.
Article written by James Blackshaw, Scientific Data Engineer at EMBL-EBI