Using genomic surveillance to track MRSA 'superbugs'
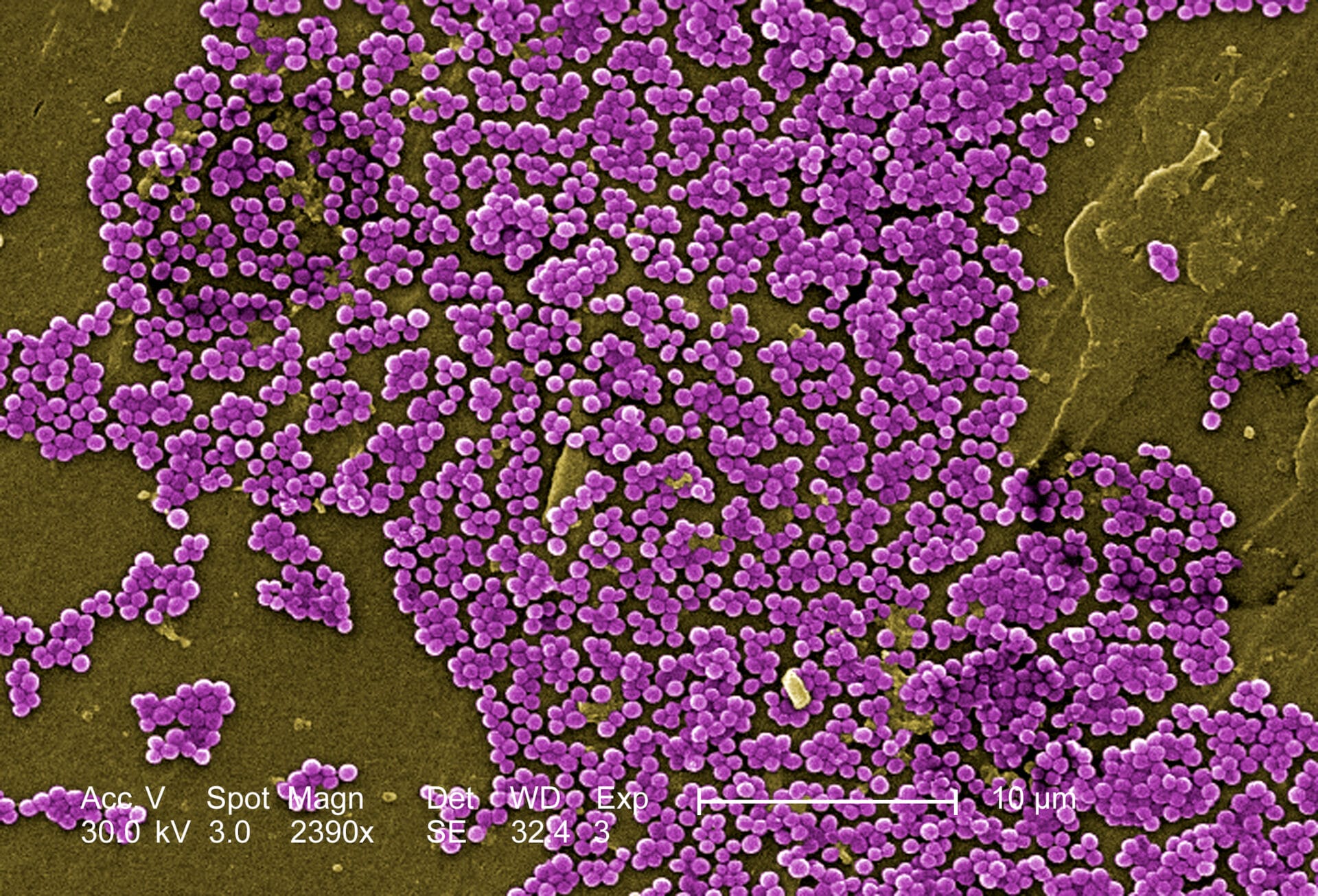
Infectious diseases can spread quickly in a hospital environment, particularly if the pathogen that causes the disease is resistant to the drugs relied on to fight it. Genomics may be able to help track these resistant pathogens and nip them in the bud before they become widespread.
- Methicillin-resistant Staphylococcus aureus (MRSA) is a type of Staphylococcus aureus that is resistant to a number of antibiotics including methicillin.
- Many of us live with S aureus and MRSA without any problems. However, MRSA can be a major challenge in hospitals – because of its antibiotic resistance, MRSA can be very difficult to treat and can spread very quickly.
- Genomics is helping to keep track of outbreaks, so that we can control them.
MRSA: The superbug
The bacterium Staphylococcus aureus (S aureus) has many faces.
Many of us happily live our whole lives with S aureus present on our skin or in our noses and experience no problems at all. But if the bacteria get further into the body, they can cause health problems. These range from mild skin infections causing redness and blisters to life-threatening infections of the heart and lungs.
Methicillin-resistant S aureus (MRSA) causes problems because it is resistant to the antibiotics that are normally used to treat these infections. This makes it much more difficult to treat – so it’s often referred to as a ‘superbug’.
MRSA infections spread quickly in contained spaces, especially in nursing homes and hospitals where people often have weaker immune systems, enabling the infection to thrive. In hospitals, MRSA can more easily enter the bodies of patients, due to cuts, wounds, and medical procedures.
Increased awareness of MRSA has enabled healthcare professionals to manage it more effectively.
Managing MRSA
An important part of managing MRSA has been gaining a deeper understanding of its biology.
In 2010, scientists at the Wellcome Sanger Institute took DNA samples from S aureus from around the world and used DNA sequencing to examine the transmission of MRSA. This was one of the first times a study like this had been done, and the results demonstrated the potential use of DNA sequencing to help reduce transmission and contain outbreaks of MRSA.
Isolating bacterial culprits
To understand what sort of bacterial infection a person has, scientists start by isolating the bacteria in a sample. This was the process used in the 2010 S aureus sequencing study.
The sampled bacteria are called ‘isolates’ and are important to ensure only one type of bacteria is sequenced at a time.
To take an isolate, a swab is used to take a sample from a patient carrying the bacteria of interest. The bacteria are then grown on a plate in the laboratory, growing in clumps known as colonies. One of the colonies of the bacteria is selected – this is the ‘isolate’ from which DNA can be extracted and sent off for sequencing.
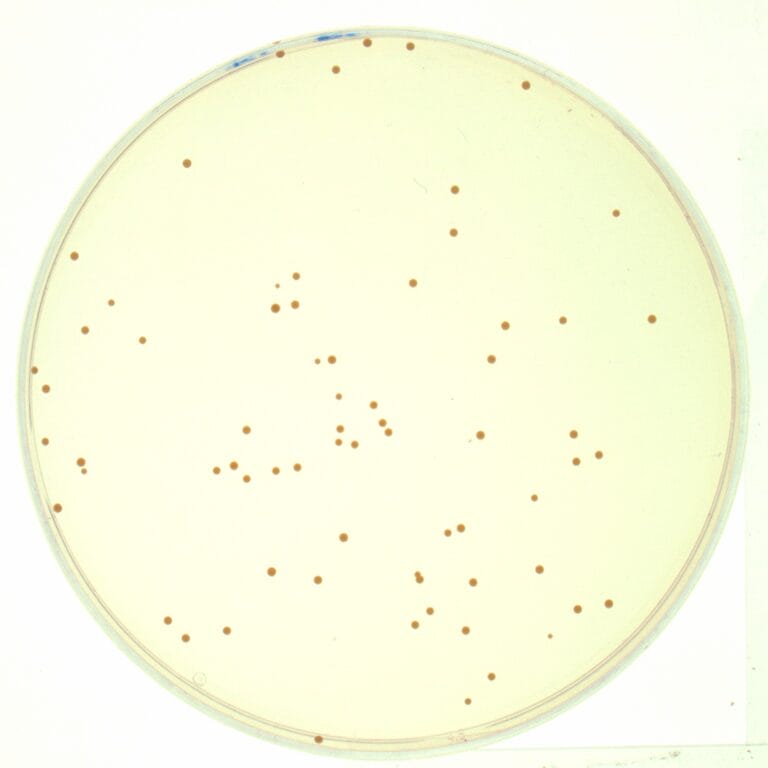
Identifying different types of bacteria with ‘typing’
For a long time, scientists used a method called ‘typing’ to work out what types of bacteria were in the isolate and help to identify the source of the outbreak. The aim is to find out whether two or more bacterial strains are related to each other and originate from the same source population (are of the same type).
One of the most common techniques for doing this is called Multilocus Sequence Typing (MLST), which involves sequencing eight genes in the bacterium’s genome. The sequence of these genes is then used to define the type of bacteria that is present in the sample. If the gene sequences are identical in two or more bacterial strains, it means that they are of the same type.
More sequence, more information
MLST has been incredibly useful, enabling scientists to separate S aureus isolates into one of 10 different subgroups based on the analysis of these eight genes.
But now, we have whole genome sequencing. This means we can determine the DNA sequence of the entire genome of an organism like S aureus in one go, which is around 2,600 genes.
Whole genome sequencing can reveal much more about different bacterial isolates than MLST’s analysis of eight genes, enabling scientists to compare detailed similarities and differences between individual S aureus isolates.
This improved level of detail means scientists can now analyse S aureus isolates and determine where in the world they cam from and how they have evolved.
The S aureus genome contains around 2,600 genes.
Using whole genome sequencing to track MRSA
The 2010 study used whole genome sequencing to analyse different MRSA samples from around the world.
Initially, they found that MRSA had been transmitted from Europe to South America and then back into Europe via Portugal. However, when they looked more closely at their data, they found an outbreak of MRSA in a UK hospital and another single case of MRSA in Denmark. The Denmark isolate was closely related to MRSA isolates found in Thailand. After further investigation, the scientists identified that the isolate found in Denmark was in fact from someone who had recently travelled from Thailand.
This demonstrated that sequencing could accurately identify the source of an individual infection and how whole genome sequencing had the potential to be used in a clinical setting. The research also suggested that by using whole genome sequencing scientists could find the source of a hospital outbreak and therefore stop it at its source before it becomes widespread.
From here, the scientists set out to demonstrate the clinical application of their findings, by looking at an outbreak of MRSA at The Rosie Hospital – part of the Cambridge University Hospitals Trust in the UK.

The 2010 study proved that whole genome sequencing could be used to track the source and evolution of MRSA strains.
How genome sequencing eradicated an outbreak of MRSA in a special care baby unit
The Rosie Hospital is home to a special care baby unit (SCBU), which cares for babies that have been born early or with a low birthweight, as well as babies that are recovering from a difficult delivery, infection or surgery.
Since these babies are very vulnerable to infection, swabs are taken on admission and then every two weeks to monitor if they come into contact with any bacteria that could cause infections.
In 2011, three babies on the SCBU at the Rosie Hospital tested positive for MRSA. Fortunately, none became unwell, but the infection prompted an investigation.

Antibiograms to determine the antibiotic resistance profile
First, the MRSA from each infected baby was tested against different antibiotics to determine their antibiotic resistance profiles – a test called an ‘antibiogram’. If samples of MRSA from different individuals are resistant to the same antibiotics, they have the same antibiotic resistance profiles and are more likely to be the same strain of bacteria.
In this case, two of the MRSA samples had the same resistance profile and one of them differed by resistance to one antibiotic. The hospital decided that the bacteria were probably linked so carried out a deep-clean of the ward and investigated all MRSA-positive swabs from the previous six months.
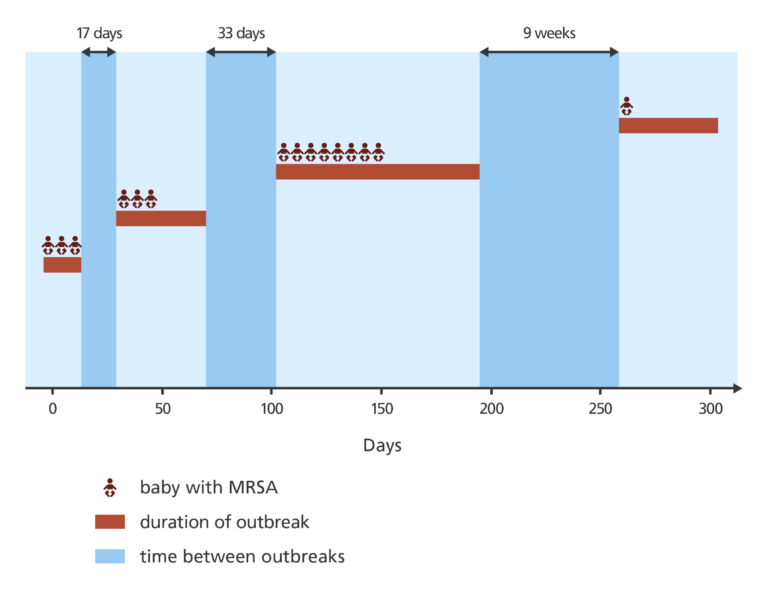
A total of 14 cases of MRSA were identified from the past six months’ samples. Nine of the cases had the same, or similar, resistance profiles to the original outbreak, and five cases had different profiles and were considered unrelated.
Genomics provides further clues
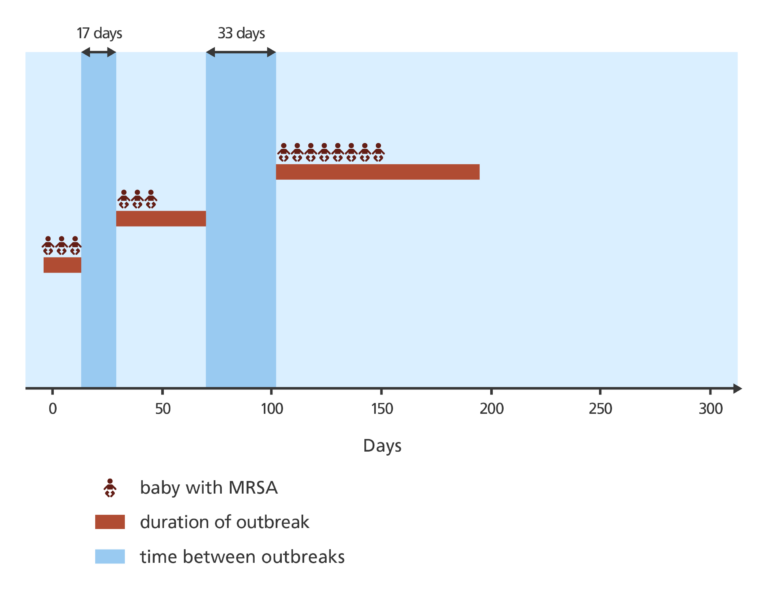
The related MRSA cases appeared in three clusters separated by 17 days and 33 days.
This pattern made it difficult to work out whether there was one single outbreak or several separate outbreaks. Normally with transmission on a hospital ward, bacterial infection is passed directly from one person to the next with no noticeable gap.
To understand more, scientists from the Wellcome Sanger Institute used DNA sequencing to explore the outbreak – and revealed more detail than the antibiograms could.
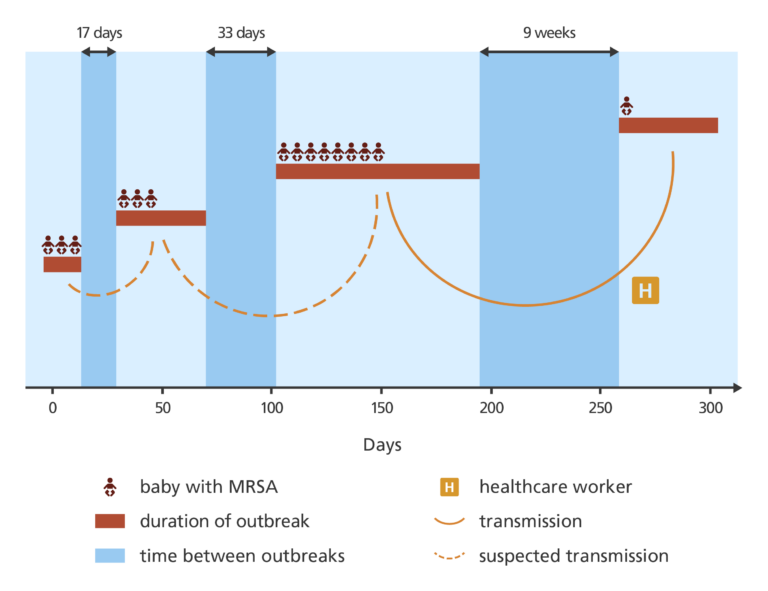
By comparing the genomes of the isolates, they found that two of the five cases considered unrelated based on their antibiograms alone were in fact related. They also confirmed that the infections were linked by a single, ongoing outbreak.
The question then was why were there gaps of many days between them? Was the outbreak being repeatedly brought into the hospital from outside, or was it originating from another ward in the hospital?

The scientists could reveal more information about the infection using whole genome sequencing than traditional antibiogram methods.
Hunting for the source of the outbreak
To find out where the cases of MRSA originated, the scientists gathered MRSA samples from other wards in the hospital, as well as GP practices and clinics in the region where patients had presented with symptoms of MRSA infection.
They then performed antibiograms on the samples to find the ones that had similar antibiotic resistance profiles to the ones identified in the SCBU. Samples with the same antibiogram then had their DNA sequenced.
When the DNA sequences of these isolates were studied, several of them were found to be closely related to the cases of MRSA on the SCBU.
Two matching MRSA samples from GP practices were from babies who had been on the SCBU ward at the same time as some of the other babies but hadn’t tested positive while they were on the ward. Some of the matching samples were taken from people related to the babies on the SCBU, including mothers who had visited their GP with abscesses (a symptom of Staphylococcal infection) who were the mothers of babies who were on the SCBU.
The scientists showed that all cases could be linked back to the SCBU – and there was no evidence that the outbreak had come from the community or other wards.
Another case of MRSA
Nine weeks later, another case of MRSA was identified on the SCBU.
It was presumed that this was a completely new outbreak of MRSA – but when the DNA was analysed it showed it to be closely related to the previous cases of MRSA on the SCBU. This suggested that the MRSA was probably being transmitted by someone working on the SCBU, but they would need evidence to confirm this.
Nipping it in the bud
To work out the source of the repeat infection, more than 100 people who worked on the SCBU provided a bacterial swab for DNA analysis – it’s common for healthcare workers to unwittingly carry MRSA, since it can frequently cause no symptoms.
Of all the samples provided by SCBU staff, one tested positive for MRSA. When DNA from that isolate was sequenced, it confirmed a link to the MRSA cases in the SCBU – suggesting the healthcare worker was the source of the most recent case on the ward.
Everyone on the ward received a series of medicated body washes to remove the MRSA. After three negative screens, the healthcare worker was considered free from MRSA and could return to work.
Finally, the outbreak was contained and eliminated.
What was the original MRSA source?
But where did the healthcare worker pick up the MRSA?
After further investigations, the MRSA strain involved in this case was found to be a strain called ST22. There are two main subtypes of MRSA ST22: one that is resistant to specific antibiotics and is commonly associated with hospitals, and one that is associated with communities.
The isolates of ST22 were found to be more closely related to the community-associated type. The most likely scenario is that one of the babies or one of the families was the original source of the MRSA infection from outside the hospital, and the healthcare worker was simply a carrier passing it between babies and families on the SCBU.
It is only through DNA sequencing that they were able to find this out!
What’s next?
This was a landmark study, demonstrating how DNA sequencing could be used in a hospital setting, and showing how sequencing of bacterial genomes can be carried out on a large scale.
Initially this study was designed to show how DNA sequencing could provide information about a historical outbreak of MRSA in a hospital. However, DNA sequencing assisted in identifying that the outbreak was ongoing and enabled the rapid identification of its origins, enabling the efficient management and containment of the outbreak.
This proves the value of DNA sequencing to help clinicians prevent the spread of MRSA and limit the number of serious infections. In serious cases, some S aureus infections can be fatal, others can require surgery to remove abscesses – but DNA sequencing could provide the insight to reduce the overall clinical burden of infectious diseases like MRSA.
Because of this study and other research over many years, Cambridge University Hospitals Trust now routinely sequences the genomes of every case of S aureus onsite to help keep one step ahead of any potential outbreaks. Not only does this help manage infection, but it can also enable scientists to identify the genetic changes that cause resistance to antibiotics and help doctors decide which antibiotics to prescribe to their patients. This technique is also being used to tackle many other pathogens including tuberculosis and gonorrhoea.
Antibiotic resistance is frequently featured in the news as a growing global problem. Gonorrhoea, for example, is becoming almost untreatable due to antibiotic resistance. Gaining a better understanding of these bacteria with genomics and DNA sequencing is providing invaluable information to help advance research at a faster rate.
