Science in the time of cholera
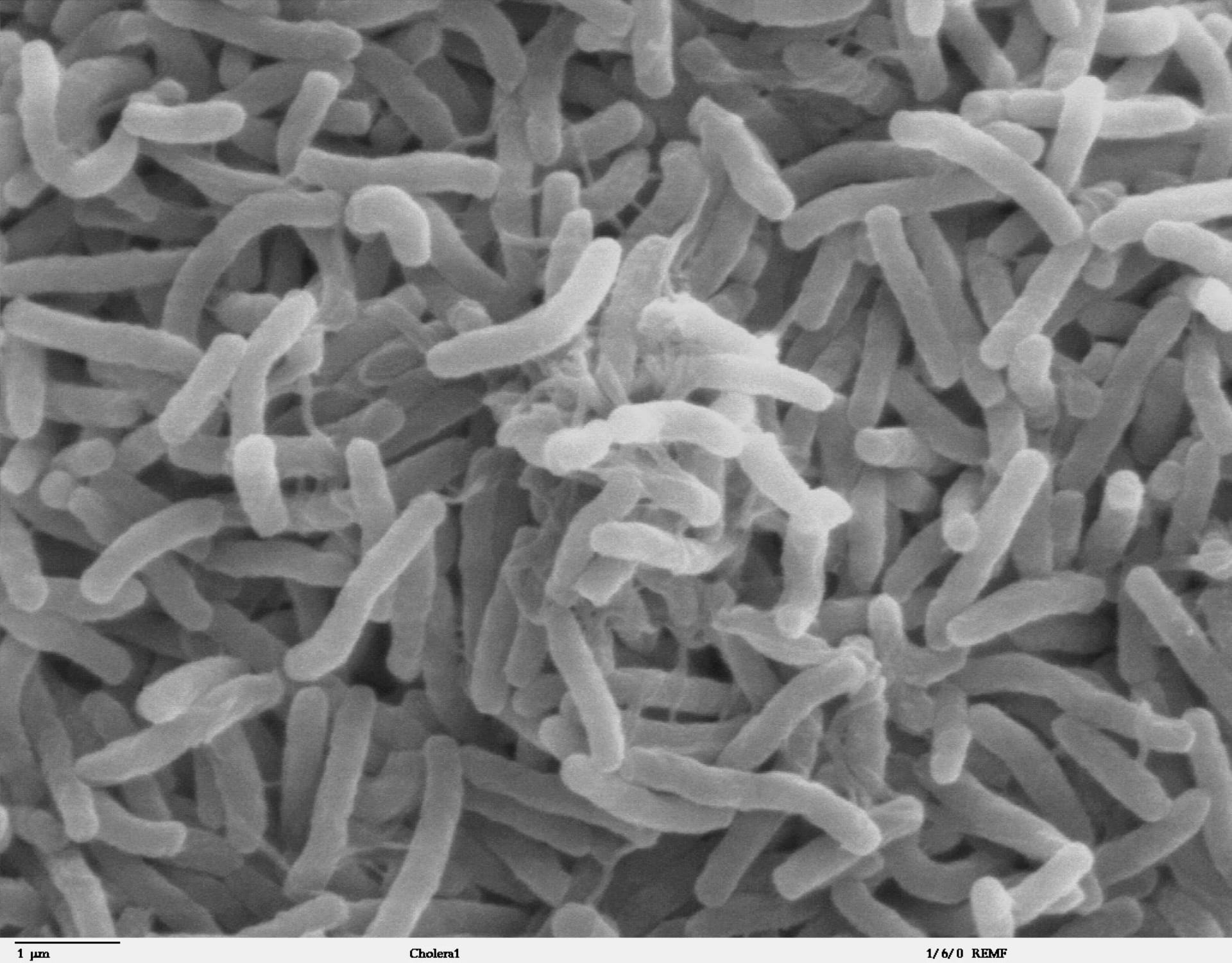
Cholera remains a major public health problem in many parts of the world, especially in places with inadequate access to clean water. Genomics research is helping to bring us closer to understanding how we may eliminate it once and for all.
Key terms
Bacteria
A group of microscopic, prokaryotic, single-celled organisms.
Genome
The complete set of genetic instructions required to build and maintain an organism.
DNA sequencing
The process of determining the order of bases in a section of DNA.
- Cholera is an extremely infectious disease of the small intestine caused by strains of the bacterium Vibrio cholerae.
- Between 1.3 and 4 million people are affected each year. Symptoms include acute diarrhoea that can last several days and cause life-threatening dehydration.
- In 2017, the World Health Organization launched a strategy to reduce cholera deaths by 90% by 2030. This includes providing safe water to prevent infection and rapid treatment of severe cases with intravenous fluids and antibiotics.
John Snow and the Broad Street water pump
The first cases of cholera in England were reported in 1831, around the same time an 18-year-old man called John Snow was completing his medical studies in London. Over the next 20 years cholera caused a series of serious epidemics, killing tens of thousands of people in England alone.
Back then, very little was known about how infectious diseases spread – or even what infectious diseases were. Many people believed that diseases like cholera and the Black Death were caused by breathing in miasma (or ‘bad air’) coming from decomposing matter.

Although John Snow had specialised in women’s health and pregnancy, he was interested in many areas of medical science, including how infectious diseases like cholera spread.
He was particularly interested in the role of water in transmitting infectious disease. In the mid-1880s, people didn’t have running water or clean ways to dispose of or treat sewage. Sewage was often dumped into open pits called ‘cesspools’ or even directly into the River Thames – before water from the river was bottled for drinking.
John Snow recognised this and suspected that sewage could be contaminating the water supply and spreading cholera – and probably many other diseases – around the city.
John Snow recognised that contaminated water could be spreading cholera.
An outbreak of cholera in Soho, London
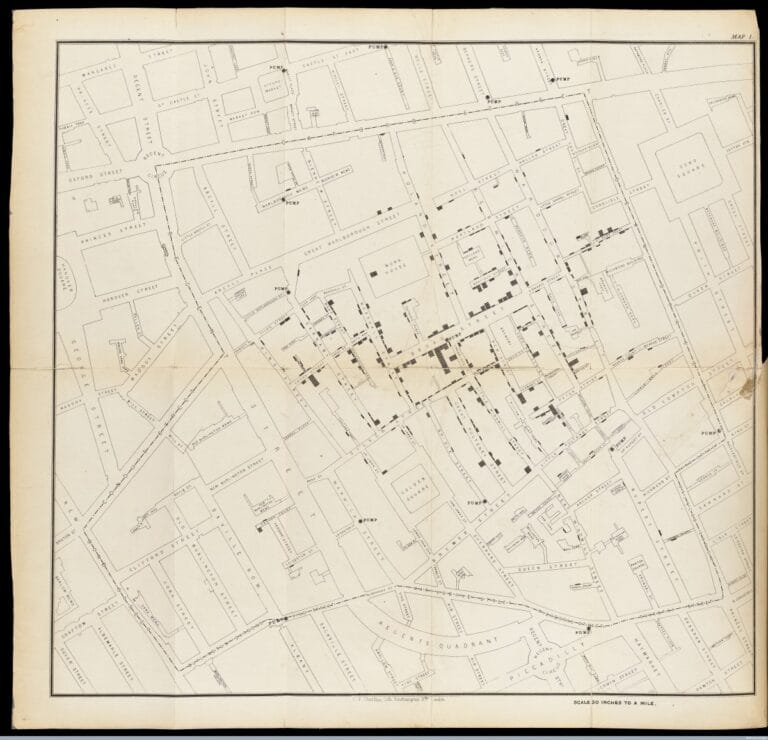
In September 1854 a particularly severe outbreak of cholera hit the Soho area of London, close to where John lived. He took the opportunity to find the source of the outbreak, working round the clock to track the infection through hospital and public records.
John constructed a map (below) showing the location of various water pumps around the city and where deaths from cholera were clustered. He showed the number of deaths at each address as a series of horizontal lines, stacked up like a pile of bodies in the street.
On 7th September 1854 John took his findings to the town officials and convinced them to take the handle off the Broad Street water pump. Although they initially refused his request, after removing the handle the outbreak of cholera almost immediately dissipated.

The German physician Robert Koch identified the cause of cholera: the bacterium Vibrio cholerae.
Identifying the cause of cholera
A few decades later, the German physician Robert Koch identified the cause of cholera: the bacterium Vibrio cholerae. Koch confirmed that the bacterium was indeed spread via unclean water or food, providing concrete support for John Snow’s theory.
John is now widely credited with establishing the field of epidemiology. To mark the importance of his discovery, the Broad Street pump is on permanent display in the London School of Hygiene and Tropical Medicine.


There are up to four million cases of cholera every year worldwide.
Cholera remains a major global health issue
Since the 1880s, many countries have built up a solid infrastructure for the distribution of clean water, as well as the disposal and treatment of sewage. This has prevented the transmission of water borne diseases such as cholera.
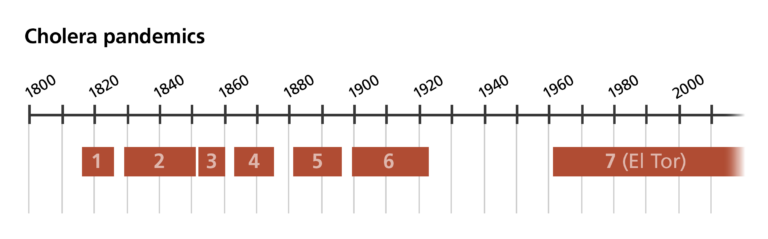
But the cholera story doesn’t end there. The disease remains a major public health issue today, with up to 4 million people affected every year. We are currently in the seventh official pandemic, which started in South Asia in 1961, reached Africa in 1971 and the Americas in 1991.
The infection is closely linked to places where people don’t have access to clean water or sewage disposal systems – such as urban slums and camps for displaced people and refugees.
For example, a devastating earthquake struck the island of Hispaniola, (which is made up of the Dominican Republic in the east and Haiti in the west. The earthquake killed more than 160,000 people and caused a loss of infrastructure to Haiti that led to political and social unrest. This loss of infrastructure contributed to a devastating cholera outbreak.
Similarly, across the early 2020s, severe floods in Pakistan, earthquakes and drought that hit Afghanistan and severe drought in Somalia have all led to increased cases of cholera within these countries.

With DNA sequencing, we can now gain a much deeper understanding of the bacterium that causes cholera by looking at its genome.
Tracking cholera outbreaks and pandemics with genomics
Six cholera pandemics have been recorded since 1816. The current, seventh pandemic is caused by a subtype of cholera called El Tor.
There are many different subtypes of cholera, but only a few that cause ‘pandemic’ disease and spread rapidly across countries and continents. Pandemic types of cholera are of primary interest for DNA sequencing because they cause the most widespread and devastating disease.
Isolating isolates
To track the spread of cholera, scientists take samples from people with the infection from different regions around the world and sequence the bacterial genomes from their samples.
The sampled bacteria are called ‘isolates’ and ensure only one type of bacteria is sequenced at a time.
By comparing the genomes of these isolates, scientists can say with confidence whether cholera bacteria from different countries are related to each other. They can also work out how closely related different isolates are and how recently they separated from each other. This can indicate how recently the isolates evolved – whether they separated from each other just a few days or weeks ago, or if they are more distantly related and separated by a few years or even decades.
This is useful information to know. If two people are found to be infected with cholera bacteria that are closely related, then it may suggest that they have both recently visited the same country or community and may even have drunk from the same contaminated water source.
That can help to track down the source, so attention can be turned to eliminating it.
Identifying outbreak origins of the 7th cholera pandemic
Sequencing a genome enables us to find out the exact order of DNA bases. Sequencing many genomes means we can compare them side-by-side to look for similarities and differences.
Scientists have done this with cholera, enabling them to draw a phylogenetic tree of cholera isolates from 1930 up to the present day. Like a family tree, a phylogenetic tree shows how different individuals are related to each other – including the single population at the root of the tree, which acts as the source of the outbreak.
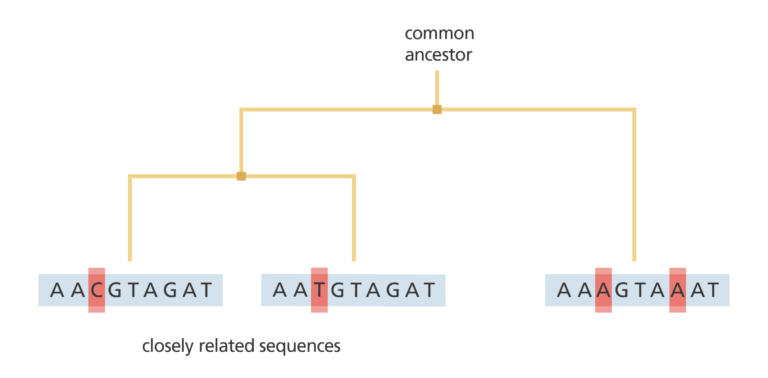
Through this study, scientists have found that all cholera outbreaks since the 1930s can be traced directly back to this source population. Each local outbreak eventually dies out – but the source population continues to exist and continues to evolve.
This asks the question: is the source population found in one place? Or does it move around?
Intriguingly, all cases of cholera from South Asia are closely related to the source population. Contrastingly, outbreaks in other parts of the world, like Africa and South America, were more distantly related to it.
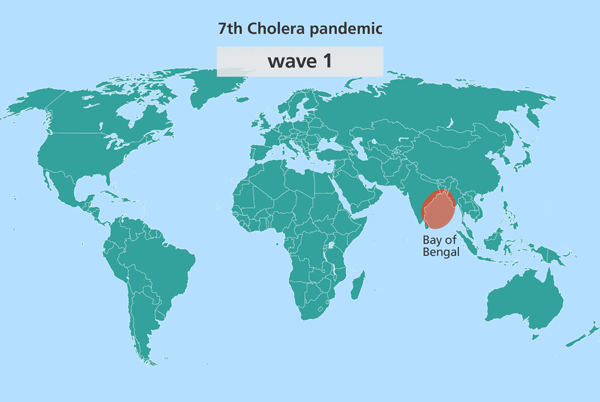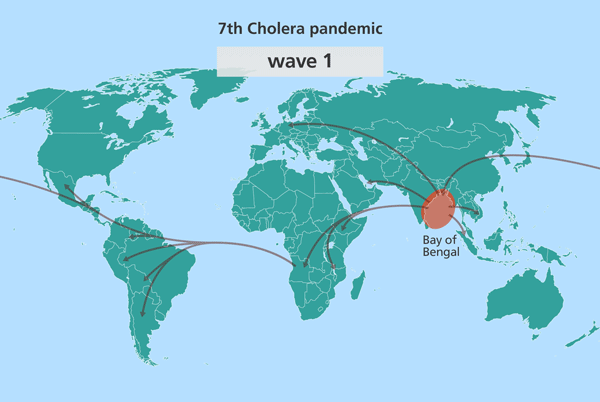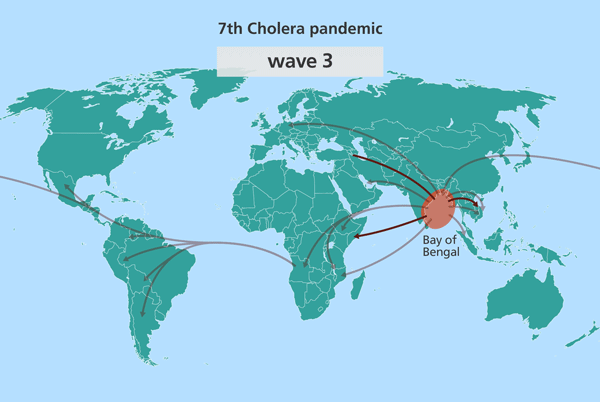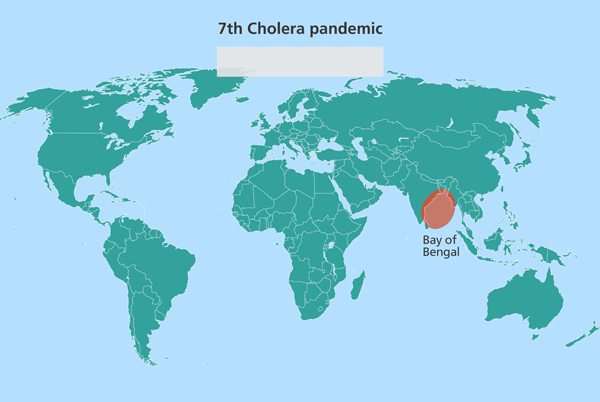
Since the isolates from South Asia are most closely related to the source population, this suggested the source might also be in South Asia. Further investigation identified this source location as being in the Bay of Bengal, in the Ganges delta, positioned in the middle of India and Sri Lanka, Bangladesh and Myanmar (Burma).
What next?
Further research has since pinpointed this stretch of water as the world’s equivalent of the Broad Street Pump identified by John Snow, from which the world’s cholera pandemics stem.
With this information, scientists could focus their efforts on tackling cholera in the Bay of Bengal and helping to prevent further spread. However, this requires resources, funding and time to understand how cholera spreads and the nature of the water sources involved.
In 2017, the World Health Organization announced ‘cholera roadmap’ – a strategy to reduce cholera deaths by 90% by the year 2030. A multifaceted approach is needed, combining genomic surveillance, surveillance, water, sanitation and hygiene, social mobilisation, treatment, and oral cholera vaccines.
Building on the foundation of John Snow’s work all those decades ago, genomics now provides us with an extremely powerful tool to make high resolution maps to show how a whole host of infectious diseases spread. These maps can be used to understand outbreaks across regions, countries and continents but also in specific local communities and hospitals. Hopefully they will enable scientists and doctors to gain the upper hand when tackling infectious diseases like cholera in the future.