Post-sequencing: storing and sharing sequenced genomes
Image credit: Greg Moss / Wellcome Sanger Institute

How are sequenced genomes stored and shared?
This is part 5 in our series looking at what happens after DNA is sequenced. You might want to start with part 1, looking at the first step of quality control.
- After a genome has been sequenced, assembled, and annotated it needs to be shared in a format that is easily and freely accessible to all.
- This can be done via a database called a genome browser.
What is a genome browser?
- Once a genome sequence has been assembled and annotated, the information needs to be stored in a database so that it can be shared with lots of people around the world.
- The visualisation of this data is done via a genome browser.
- Making data openly accessible and easily visualised in this way is hugely important in helping to support and progress scientific research across the globe.
- The initial aim of having a genome browser was to display the human genome sequence and provide a platform to allow people to ‘browse’ and analyse the DNA sequence for themselves.
- Different genome browsers collaborate together to share data and ideas to ensure that they are presenting the data in a consistent and uniform way.
- The information on many browsers can be viewed for free and by anyone anywhere without restriction. The only prerequisite is an internet connection!
Examples of genome browsers include:
- Ensembl, a joint project between the European Bioinformatics Institute (EBI), part of the European Molecular Biology Laboratory (EMBL), and the Wellcome Trust Sanger Institute in the UK. Ensembl launched in 1999, just before the release of the first draft of the human genome by the Human Genome Project.
- UCSC, a genome browser based at the University of California Santa Cruz, on the west coast of the USA, which launched in 2001, initially to showcase the draft sequence of the human genome.
- National Center for Biotechnology Information’s (NCBI) map viewer, based in Maryland in the USA, provides a wide variety of data taken from genome mapping and sequencing data.
What data is available and how does it look on the browser?
- Genome browsers such as Ensembl and UCSC use coloured and interactive graphics to present the complex data in an integrated manner.
- The basic structure of the display on many genome browsers is to show the genome sequence horizontally across the screen with certain elements presented in specific colours and shapes according to a key.
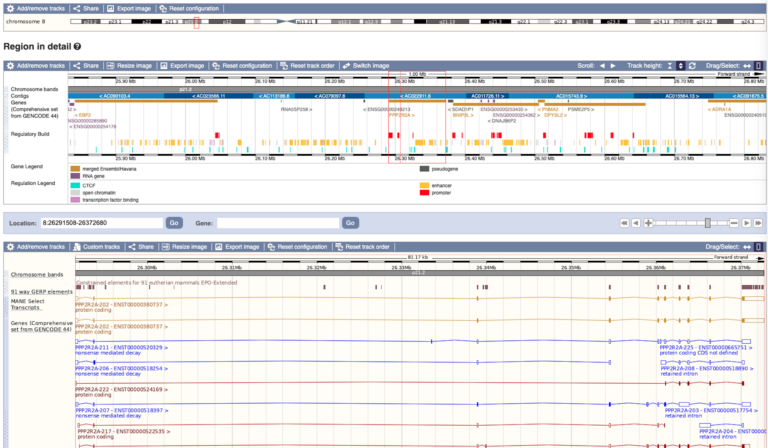
- On Ensembl, each species has its own page on the browser, which enables you to view and download the DNA sequence and explore other, more specific, information about that genome.
- The data is held in data ‘tracks’ which can be switched on and off depending on what aspect of the genome you want to look at. For example, you can view protein alignments, genetic variants, genes and much more.
- To find a specific gene or region of the genome, the user can type in the name of the gene (for example, BRCA1) or a particular position on a chromosome using genomic coordinates (e.g. 17:43044295-43125483). The user will then be able to explore the specific data for that region and customise the view if they wish.
- As well as analysing a gene within a single species and getting information about it, genome browsers also allow you to compare genes and genomic regions across different species.
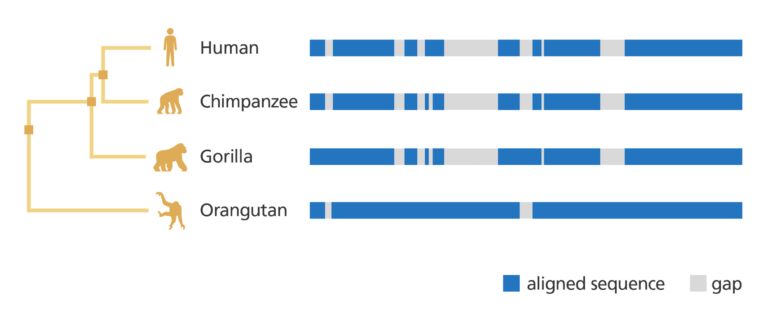
- You can explore genetic variation and find out where genetic variants associated with specific diseases or characteristics are located on the chromosomes.
- From the overview of a chromosome you can zoom in to find the sequence of bases in the DNA (A, T, C and G).
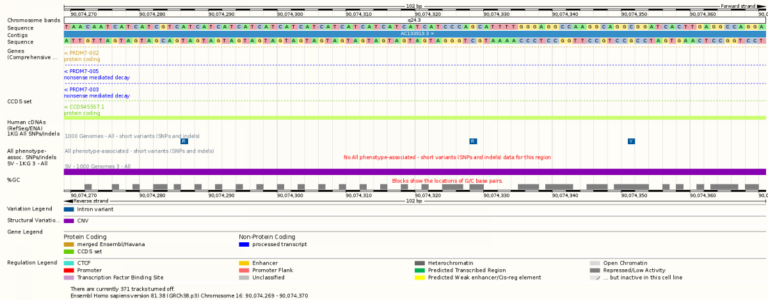
- The amount of genomic data available depends on the species you are looking at. For example, the human is the species for which the most data is available as a lot of scientists are working with human genomic data. In contrast there is currently very little genomic information on the aardvark.
How often is the data updated?
- Ensembl release a new version of their database every two to three months as part of their ‘release cycle’ (you can read more about it here)
- As well as data updates, genome browsers may also release new tools and ways of visualising the data.
- When updates are made, the old versions are still available to view in their archives.
- Genome browsers like Ensembl and UCSC are constantly evolving. The data on the browsers is always changing so it is important for those using them to keep up to date with the latest version.
- Ensembl ensures the browser remains a user-friendly, intuitive and reliable interface.
Who uses a genome browser?
- Some prior knowledge of genetics and molecular biology is needed to use a genome browser, to understand what the data is about and what the interface displays.
- Genome browsers, such as Ensembl, are used by scientists from different groups:
- Wet-lab scientists – lab scientists working directly with biological material.
- Bioinformaticians – computer scientists who create mathematical models and sophisticated computer programmes for gathering, viewing and analysing biological data such as genetic/genomic data.
- Clinicians – who examine the human genome to help with the development of new tools for diagnosing disease.
- Teachers – who use the browsers to support their teaching of molecular biology and genetics in the classroom.
Where does the data come from?
- The data on genome browsers is collected from collaborations with various research projects and databases such as the International Nucleotide Sequence Database Collaboration (INSDC), Single Nucleotide Polymorphism database (dbSNP), the Encyclopedia of DNA Elements (ENCODE), and 1000 Genomes Project.
- INSDC is a collaboration between the European Bioinformatics Institute (EMBL-EBI), NCBI and the DNA Data Bank of Japan (DDBJ). This is where genome browsers such as Ensembl get the raw genomic sequence.
- dbSNP is a free, public, online archive developed and hosted by the National Center of Biotechnology Information in the US. Its goal is to act as a one-stop-shop database for genetic variants (SNPs and a whole range of other variations) in any organism.
- ENCODE is a research project that launched as a follow-up to the Human Genome Project. It aimed to identify and characterise all the functional parts of the human genome to reveal how it works.
- 1000 Genomes Project was the first project to aim to sequence the genomes of a large number of people to produce a detailed catalogue of human genetic variation.
- Before being loaded on to the database and visualised in genome browsers, the data is crunched and sorted so that it can be presented in a more user-friendly way for people viewing the information.
How do we get from genome sequence to genome browser?
- Once the assembly is publicly available, the genome browser can start its annotation using ‘biological evidence’ so that genes and transcripts can be identified.
- This biological evidence comes in the form of nucleotide and amino acid sequences, all of which need to be put into the context of all of the other genetic information we have for that organism .
- The first step in doing this is to map and align the biological evidence against the reference genome (assembly) for that organism.
- Software is then run to identify where the sequences map to the genome and allowing gene structures with exons and introns to be drawn and displayed on the browser.

- This can then be combined with information about what those genes do, what diseases or characteristics they are associated with and other genomic information.
- The underlying data also undergoes quality checks and assessments on a regular basis.
- Performing these quality checks may sometimes cause a delay in the uploading of new data to the database and visualisation on the genome browser. However, it is an imperative step to ensure that the quality of the data remains of a high standard for downstream analyses by the user communities.