Genomic surveillance in action

Genomic surveillance sequences and monitors the genetic material of disease-causing pathogens, such as viruses and bacteria, so we can identify, track and control them. It’s become an essential tool in tackling disease epidemics.
Key terms
DNA sequencing
The process of determining the order of bases in a section of DNA.
Mutation
A change in the DNA sequence, which may have positive, negative or neutral effects on the organism.
Immunity
An organism’s ability to resist an infection or disease.
Vaccine
A type of treatment that provides active acquired immunity against a specific pathogen.
- Genomic surveillance uses modern DNA sequencing methods to track pathogens as they evolve.
- Samples are mapped geographically and through time to monitor how their genome changes.
- This information can help governments and health agencies to adapt to new health threats, or to existing diseases that become harder to diagnose or to treat.
- Genomic surveillance is also very important for the creation of new vaccines.
Genomic surveillance in action
Here are three short case studies demonstrating genomic surveillance in action.
Each showcases how our ability to sequence genomes and track variants of pathogens is revolutionising the way we understand and manage outbreaks of serious disease.
1. Covid-19
Genomic surveillance is very effective at tracking virus outbreaks – but is only as good as the number of samples analysed and the width of the area surveyed.
Genomic surveillance works extremely well when tracking virus outbreaks.
This is because virus DNA or RNA can change or mutate during replication, just like that of all living creatures – although it’s debatable whether viruses are actually ‘alive’.
In the case of SARS-CoV-2 – the virus that causes Covid-19 – genomic surveillance has allowed us to monitor how it changes over time and evolves into the different variants we see on the news. We can also monitor genetic changes that occur frequently throughout the virus’s evolution – such as the E484K mutation seen several times in different variants.
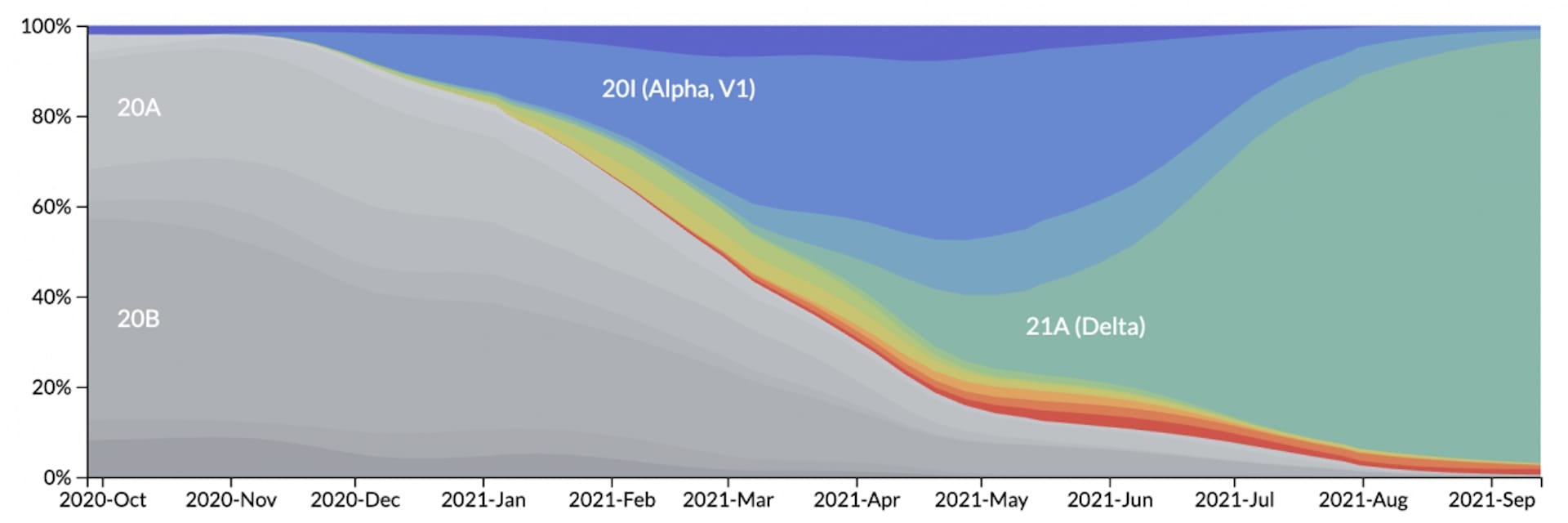
The use of genomic surveillance dramatically increased during the Covid-19 pandemic, enabling scientists and governments around the world to determine which variants are present in their country.
It can also help determine how infectious or dangerous new variants are, by monitoring how quickly they spread and how often they are found in people who required treatment or have died.
By monitoring the SARS-CoV-2 genome, we can also see if any new variants are able to evade the protection provided by previous infections or vaccines. This shows us if people become infected repeatedly and, if so, by which variant. If the same variant is constantly able to infect people who are fully vaccinated, or have had a previously confirmed infection, then it would be classed as an escape variant.
The potential and limits of genomic surveillance
While the Covid-19 pandemic showcased the power and potential of genomic surveillance, it also displayed its biggest weakness: it’s only as good as the number of samples analysed and the width of the area surveyed.
Some countries can carry out a lot of genomic surveillance, while others have fewer facilities. This means there may be many more variants than we realise.
In 2022, the World Health Organization developed a new strategy to improve the way genomic surveillance is done around the world, so we can respond quickly to future pathogens that have the potential to cause an epidemic or pandemic.

2. Ebola
Genomic surveillance has provided great help in tracking and halting Ebola outbreaks.
Ebola is a deadly disease caused by the Ebola virus. It spreads rapidly through contact with body fluids, and up to 90% of people infected with the virus will die within days.
The Ebola virus is usually found in Central and West African countries. In recent decades, it has sprung up sporadically, with little warning. This includes an outbreak between 2013 and 2016 in Guinea, Liberia and Sierra Leone, and later spreading to other countries, with more than 28,000 reported cases.
Genomic surveillance has provided great help in tracking and halting these outbreaks. Sequencing the viral genome from people with Ebola and comparing it to previous outbreaks has helped to determine patterns of transmission.
For example, if the viral genome from a new patient was very similar to others’, it was probably from a known source – and so unlikely to be a new outbreak. But if the viral genome had many differences, there was likely a new outbreak of the disease.
This was especially important when someone fell ill in an area outside of the main epidemic area: could their virus be traced back to the main epidemic? Or were they the first person affected by a new outbreak?
Understanding how infection can lie dormant
The genomic surveillance performed during the 2013-16 West African Ebola epidemic became unexpectedly useful in 2021, when a new cluster of cases emerged in Guinea. Their genetic sequence was almost identical to that of the earlier epidemic.
This led to speculation as to whether the virus lay dormant in a survivor before becoming infectious again years later – opening new areas of research about how the virus behaves.

3. MRSA
Genomic surveillance can also provide vital information for outbreaks on a far smaller scale.
Epidemics and pandemics aren’t the only times that genomic surveillance is useful. It can also provide vital information for outbreaks on a far smaller scale, as one English hospital found out.
During an outbreak of the superbug MRSA (the bacteria Methicillin-Resistant Staphylococcus aureus) in the hospital’s maternity unit, genomic surveillance determined that most cases came from a common source.
Despite proper actions to deep-clean the wards and implement changes, a second outbreak occurred – and this was linked to the first by sequencing.
After further investigations, 3 members of staff were found to be carrying symptomless MRSA. Sequencing data linked the genome from one of the staff to the MRSA found on the babies.
Treating and decolonising the affected staff members broke the transmission pathway, ending the cycle of outbreaks and making the maternity ward safe again.

What about genomic surveillance in the future?
As we become able to sequence genomes even faster and cheaper, the potential of genomic surveillance continues to grow.
The greater the number of genomes we can sequence and compare, the more closely we will be able to monitor outbreaks of disease and understand how they spread, mutate and respond to medicines or vaccines.